
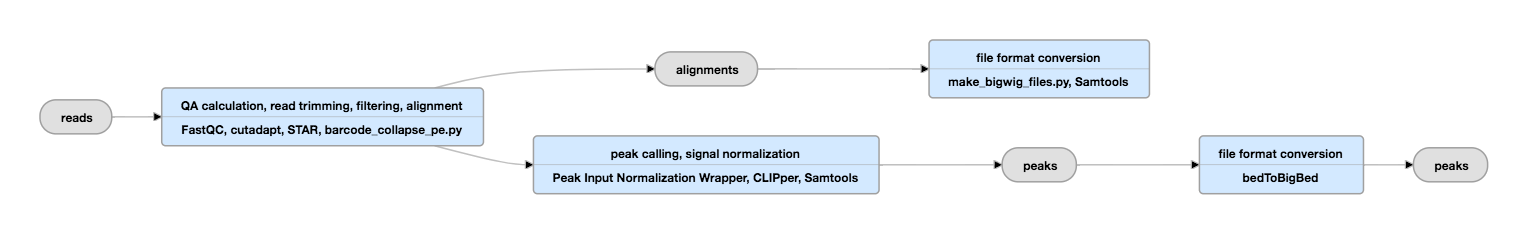
Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. Same scale is used in all panels in order to allow comparisons of the evaluated binding sites. (d) Region with the highest number of CLIP cDNA deletions. (c) Region with the second highest iCLIP cDNA count. This region also has the highest YCAY score. (b) Region with the highest iCLIP cDNA count.

The YCAY score track shows the YCAY score at each position, while the YCAYs track shows the position of YCAY motifs. The iCLIP truncations track shows the position of iCLIP cDNA truncations (FDR < 0.05), with peak height corresponding to the cDNA counts. The CLIP deletions track shows low FDR deletion sites (FDR < 0.001, the positions were re-defined), with peak height corresponding to the number of sequences containing deletions at the sites. In each panel, the CLIP cDNAs track shows the cluster of CLIP cDNAs without deletions. iCLIP cDNA deletion sites were re-defined as described in Materials and methods.Ĭomparison of CLIP and iCLIP analysis of Nova binding to its primary RNA target, the Meg3 non-coding RNA. The number of cDNAs was determined by considering the random barcode. (d) Occurrence of iCLIP cDNA truncations (black), deletions (blue) or truncations of cDNA with deletions (orange) around the re-defined CLIP deletion sites. The red line shows the starting position of YCAY motifs, and the green line shows the starting position of TTTCAY motifs. (c) Similar to (b), but the occurrence of YCAY motifs starting closest to redefined CLIP cDNA deletions, where the position of deletions mapping within TTT motifs is assigned to the middle of TTT. (b) The occurrence of YCAY motifs starting closest to the iCLIP cDNA truncations (FDR < 0.05). The dashed line shows the background occurrence of YCAY motifs.

#Clip eclip iclip plus
The black line shows the starting position of YCAY motifs on all cDNAs, whereas the light blue and yellow lines show the starting position on the plus or minus strand of the genome. (a) The occurrence of YCAY motifs around the CLIP cDNA deletions (FDR < 0.001). ICLIP cDNA truncations identify RNA cross-link sites of Nova proteins with nucleotide resolution.


 0 kommentar(er)
0 kommentar(er)
